NeurIPS Poster, ‘21,
PhysChem: Deep Molecular Representation Learning via Fusing Physical and Chemical Information
Summary
- Used physicist network (PhysNet) and chemist network (ChemNet) simultaneously, and each network shares information to solve individual tasks.
- PhysNet: Neural physical engine. Mimics molecular dynamics to predict conformation.
- ChemNet: Message passing network for chemical & biomedical property prediction.
- Molecule without 3D conformation can be inferred during test time.
Preliminaries
-
Molecular representation learning:
Embedding molecules into latent space for downstream tasks. -
Neural Physical Engines
Neural networks are capable of learning annotated potentials and forces in particle systems.
HamNet proposed a neural physical engine that operated on a generalized space, where positions and momentums of atoms were defined as high-dimensional vectors. -
Multi-task learning
Sharing representations for different but related tasks. -
Model fusion
Merging different models on identical tasks to improve performance.
Notation
Graph $\mathcal{M} = (\mathcal{V}, \mathcal{E}, n, m, \mathbf{X}^v, \mathbf{X}^e)$
- $\mathcal{V}$: set of $n$ atoms
- $\mathcal{E}$: set of $m$ chemical bonds
- $\mathbf{X}^v \in \mathbb{R}^{n \times d_v} = (x^v_1, …, x^v_n)^\top$: matrix of atomic features
- $\mathbf{X}^e \in \mathbb{R}^{m \times d_e} = (x^e_1, …, x^e_m)^\top$: matrix of bond features
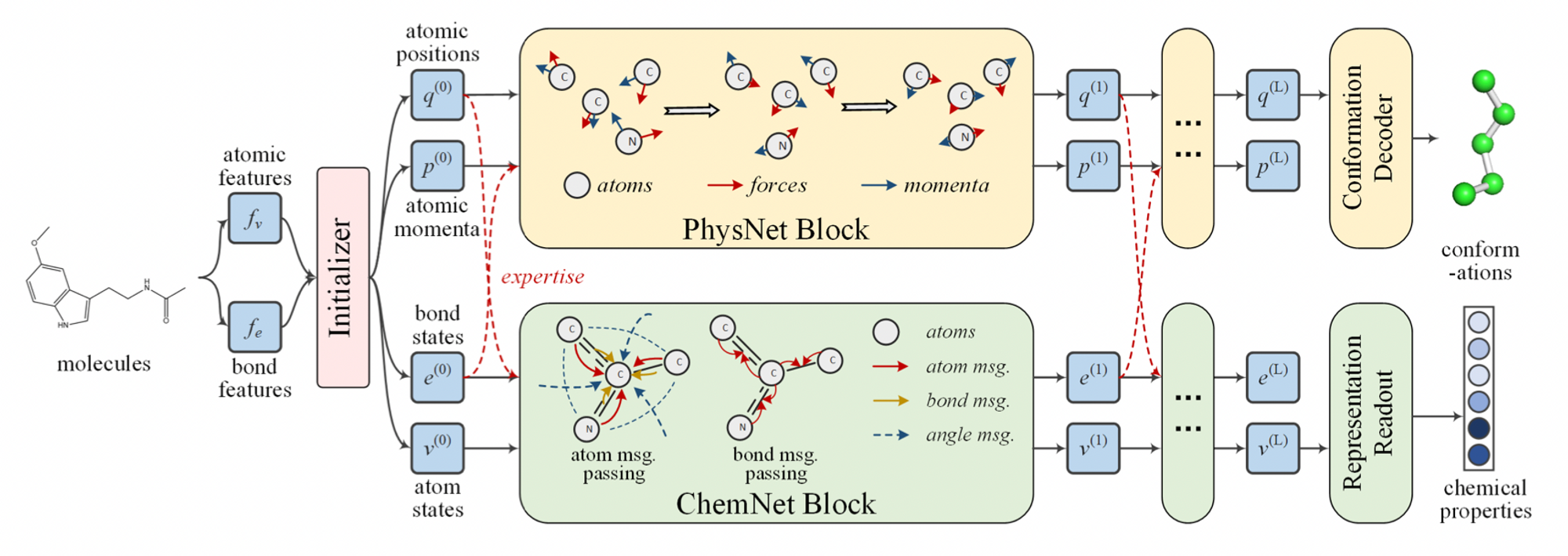
Model
 Figure 1. PhysChem Architecture
Figure 1. PhysChem Architecture
-
Initializer
- Input: atomic features, bond features (from RDKit)
- Layer: fully connected layers
- Output:
- bond states, atom states for ChemNet
$v^{(0)}_i = \text{FC}(x^v_i), i\in \mathcal{V}$
$e^{(0)}_{i,j} = \text{FC}(x^e_{i,j}), (i, j)\in \mathcal{E}$ - atom positions, atomic momenta for PhysNet
Bond strength adjacency matrix
$$A(i,j)=\begin{cases}0, & \text{if $(i,j) \notin \mathcal{E}$} \\ \text{FC}_{\text{sigmoid}}(x^e_{i,j}), & \text{if $(i,j) \in \mathcal{E}$} \end{cases}$$ $\tilde{V} = \text{GCN}(A, V^{(0)})$
${ (q^{(0)}_i \oplus p^{(0)}_i)} = \text{LSTM}({\tilde{v}_i}), i \in \mathcal{V}$
-
PhysNet
- PhysNet is inspired by HamNet.
HamNet showed that neural networks can simulate molecular dynamics for conformation prediction. - Directly parameterize the forces between each pair of atoms.
- Consider the effects of chemical interactions(e.g. bond types) by cooperating with ChemNet’s bond states.
- Introduces torsion forces.
- Output: 3D conformation
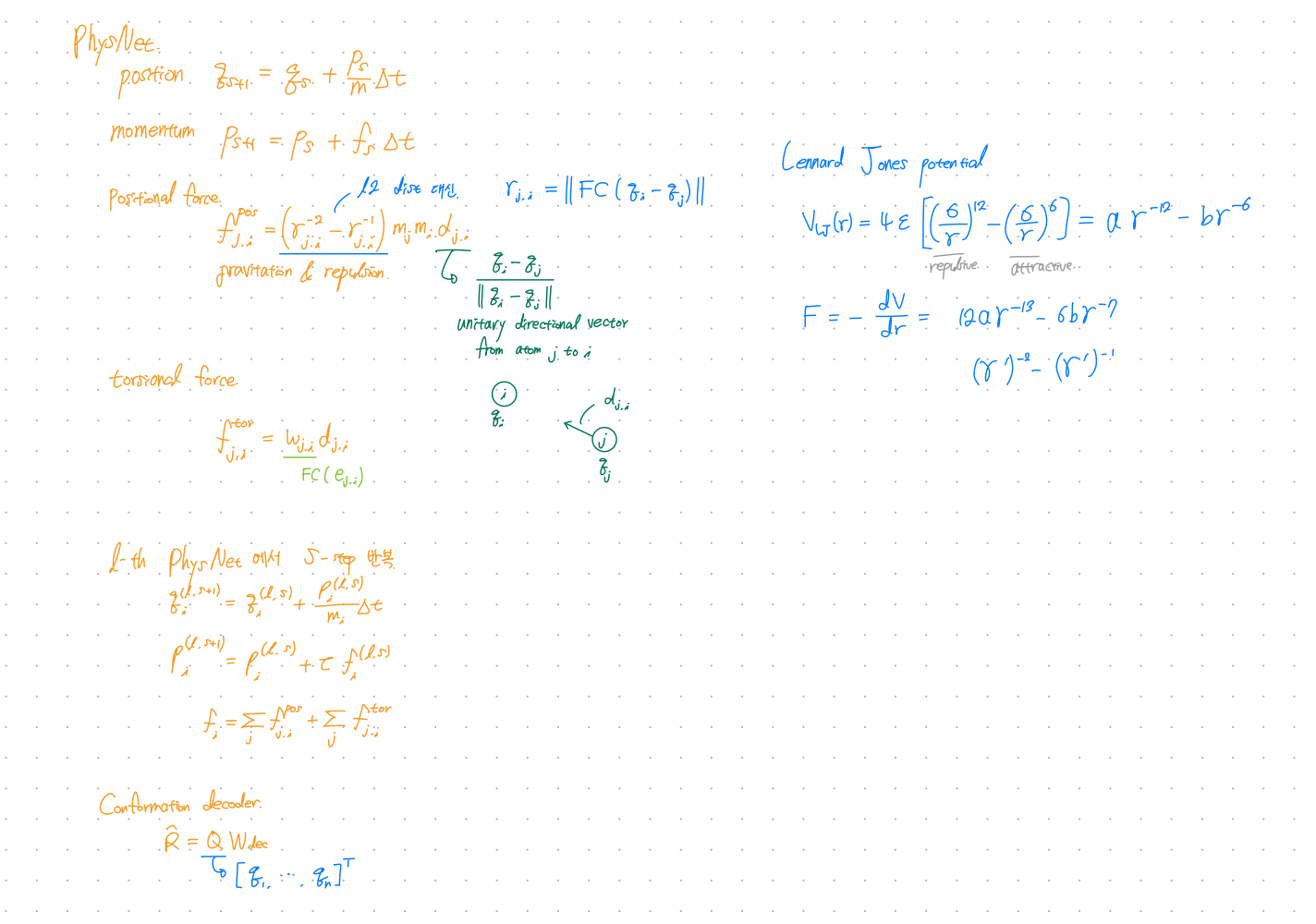
- PhysNet is inspired by HamNet.
-
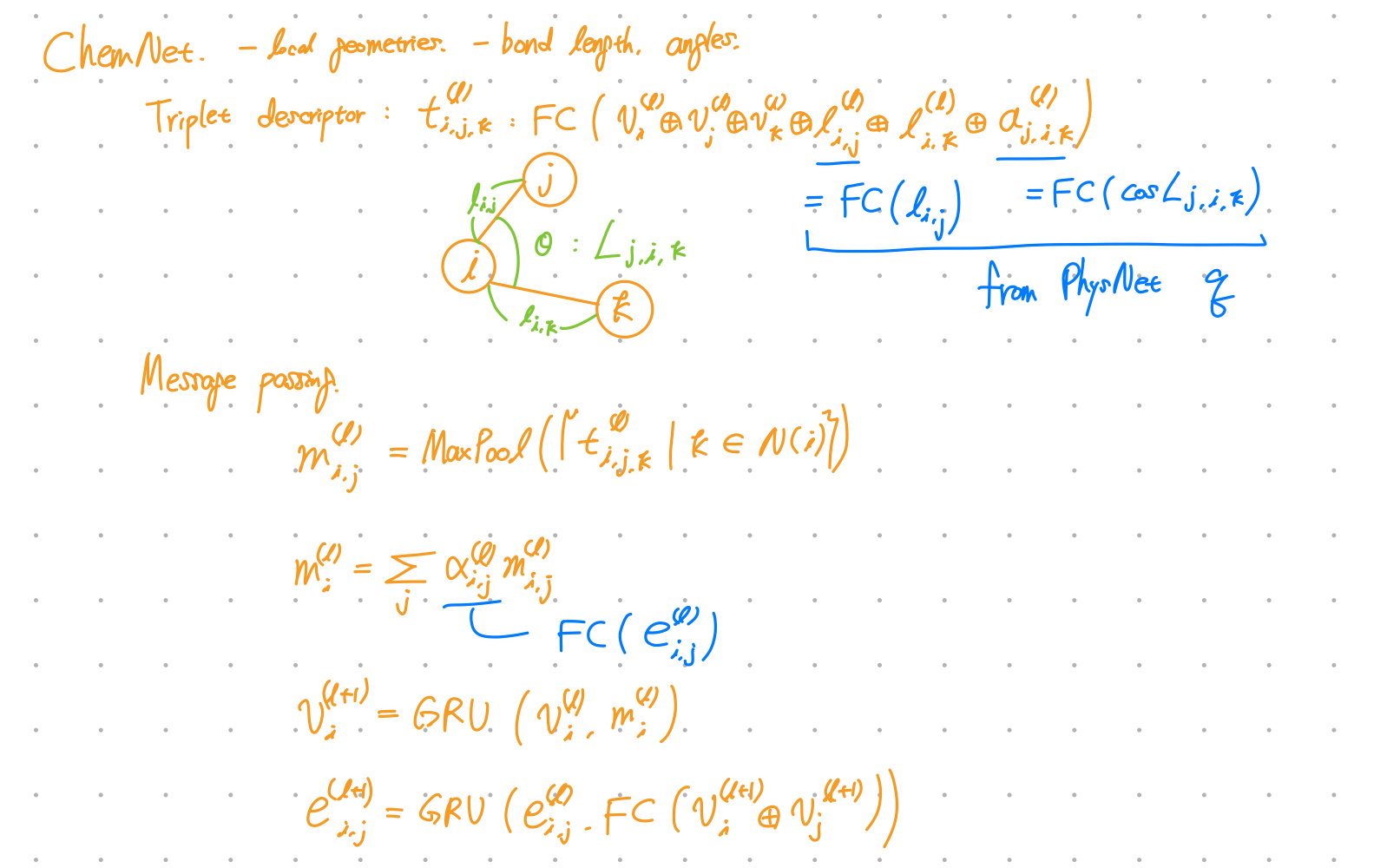
ChemNet
- ChemNet modifies MPNN(message passing neural network) for molecular representation learning.
- Output: Molecule representation
Loss
-
$L_{\text{phys}}$: Conn-k loss for Conformation prediction (PhysNet)
$k$-hop connectivity loss
$L_{\text{Conn}-k}(\hat{\mathbf{R}}, \mathbf{R}) = |\frac{1}{n} \hat{\mathbf{C}}^{(k)} \odot (\hat{\mathbf{D}} - \mathbf{D}) \odot (\hat{\mathbf{D}} - \mathbf{D}) |_{F}$
$\odot$: element-wise product
$| \cdot |$: Frobenius norm
$(\hat{\mathbf{D}} - \mathbf{D})$ : distance matrix of the real and predicted conformations $(\hat{\mathbf{R}} - \mathbf{R})$
$\hat{\mathbf{C}}^{(k)}$: normalized $k$-hop connectivity matrix
-
$L_{\text{chem}}$: MAE or Cross entropy loss for Property prediction (ChemNet)
-
Total loss
$L_{\text{total}} = \lambda L_{\text{phys}} + L_{\text{chem}}$
Checkpoints
-
Is Conn-k loss generally used in other conformation prediction models?
No! But seems related to local distance loss.
-
Is triplet descriptor generally used in other models?
No!